
As a first investigation into the existence of co-morbidities in post-medieval plague victims we applied a non-targeted pathogen screening approach offered through the HOPS platform 17 to skeletal material from a suspected 15 th–16 th century plague burial from Vilnius, Lithuania. The influence of co-infections as exacerbating the progression of certain diseases is an established phenomenon 13, 14 and has been proposed to partially account for plague’s unusually high mortality in the past 15, 16, its potential influence on selective mortality 16, as well as its persistence and subsequent rapid extinction in post-medieval Europe 8. Here we present the application of an ancient pathogen detection method to identify co-morbidities in archaeological specimens associated with an historical epidemic. Though demonstration of these techniques is currently restricted to single microbial organisms, they indeed offer the flexibility to detect co-morbidities on historical timescales, thus giving a glimpse into past disease ecology. More recent approaches have explored the analytical resolution of broad multi-species enrichments 10 or fully non-targeted approaches 11, 12 for pathogen detection. By design, this process requires a priori knowledge of a taxon of interest, which is typically acquired through either initial molecular screening 7, inference from historical context 8, diagnostic gross pathology 9, or combinations thereof 5. Genomic investigations of ancient microbes have largely been made possible via selective enrichment for DNA of a target organism. Genome-level analyses of ancient bacteria and viruses have revealed unexpected patterns of pathogen dispersal in the past, and have prompted discussants to revise long standing theories about the evolution of some of our most well-studied diseases such as plague 4, tuberculosis 5 and smallpox 6 to name but a few.
Increased sequencing capacity made available by technological innovations has translated into concomitant increases in temporal depth for human genomes 1 as well as those from commensal 2 and invasive 3 microbes. Through this we offer an alternative hypothesis for the history and evolution of the treponemal diseases, and posit that yaws be considered an important contributor to the sudden epidemic of late 15 th century Europe that is widely ascribed to syphilis.Īncient DNA analyses have the potential to reveal history that is hidden within archaeological samples.

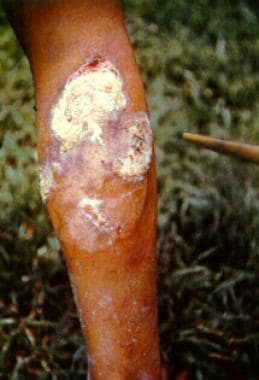
Our finding in northern Europe of a disease that is currently restricted to equatorial regions is interpreted within an historical framework of intercontinental trade and potential disease movements. In addition to Yersinia pestis, we detected and genomically characterized a septic infection of Treponema pallidum pertenue, a subtype of the treponemal disease family recognised as the cause of the tropical disease yaws. Our application of a non-targeted molecular screening tool for the parallel detection of pathogens in historical plague victims from post-medieval Lithuania revealed the presence of more than one active disease in one individual.
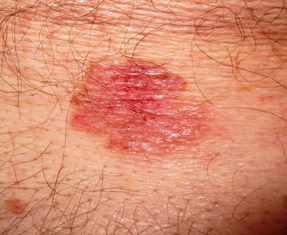
Developments in techniques for identification of pathogen DNA in archaeological samples can expand our resolution of disease detection.


 0 kommentar(er)
0 kommentar(er)
